Welcome to DeepProg’s documentation!
- Installation
- Tutorial: Simple DeepProg model
- Tutorial: Ensemble of DeepProg model
- Tutorial: Advanced usage of DeepProg model
- Tutorial: use DeepProg from the docker image
- Case study: Analyzing TCGA HCC dataset
- Tutorial: Tuning DeepProg
- License
- simdeep package
- Submodules
- simdeep.config module
- simdeep.coxph_from_r module
- simdeep.deepmodel_base module
- simdeep.extract_data module
- simdeep.plot_utils module
- simdeep.simdeep_analysis module
- simdeep.simdeep_boosting module
- simdeep.simdeep_distributed module
- simdeep.simdeep_multiple_dataset module
- simdeep.simdeep_utils module
- simdeep.survival_utils module
- Module contents
Introduction
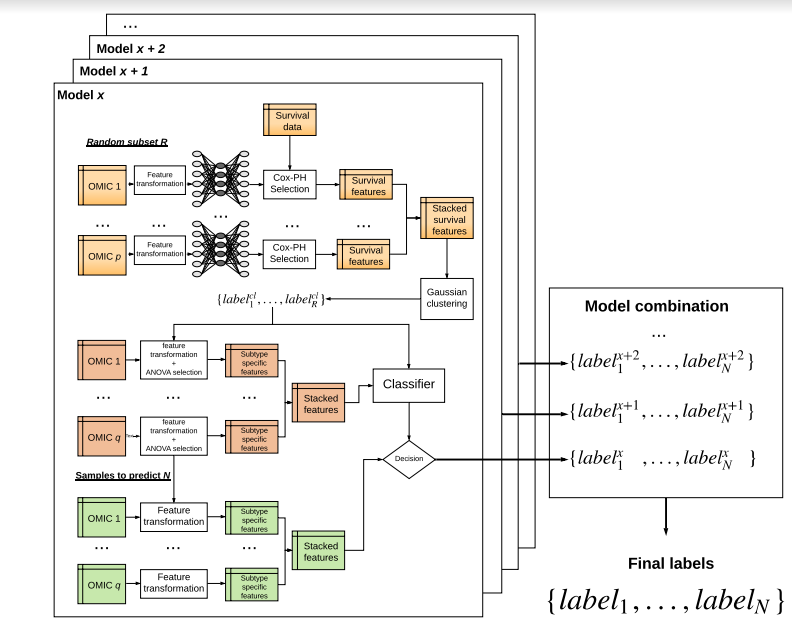
This package allows to combine multi-omics data for individual samples together with survival. Using autoencoders (default) or any alternative embedding methods, the pipeline creates new set of features and identifies those linked with survival. In a second time, the samples are clustered with different possible strategies to obtain robust subtypes linked to survival. The robustness of the obtained subtypes can then be tested externally on one or multiple validation datasets and/or the out-of-bags samples. The omic data used in the original study are RNA-Seq, MiR and Methylation. However, this approach can be extended to any combination of omic data. The current package contains the omic data used in the study and a copy of the model computed. However, it is easy to recreate a new model from scratch using any combination of omic data. The omic data and the survival files should be in tsv (Tabular Separated Values) format and examples are provided. The deep-learning framework to produce the autoencoders uses Keras with either Theano / tensorflow/ CNTK as background.
Access
The package is accessible at this link: https://github.com/lanagarmire/DeepProg.
Contribute
Issue Tracker: github.com/lanagarmire/DeepProg/issues
Source Code: github.com/lanagarmire/DeepProg
Support
If you are having issues, please let us know. You can reach us using the following email address:
Olivier Poirion, Ph.D. o.poirion@gmail.com
Data availability
The matrices and the survival data used to compute the models are available here: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00930-x/
Citation
This package refers to our study published in Genome Biology: Multi-omics-based pan-cancer prognosis prediction using an ensemble of deep-learning and machine-learning models, https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00930-x
License
The project is licensed under the MIT license.